Pathogens, Free Full-Text
Por um escritor misterioso
Descrição
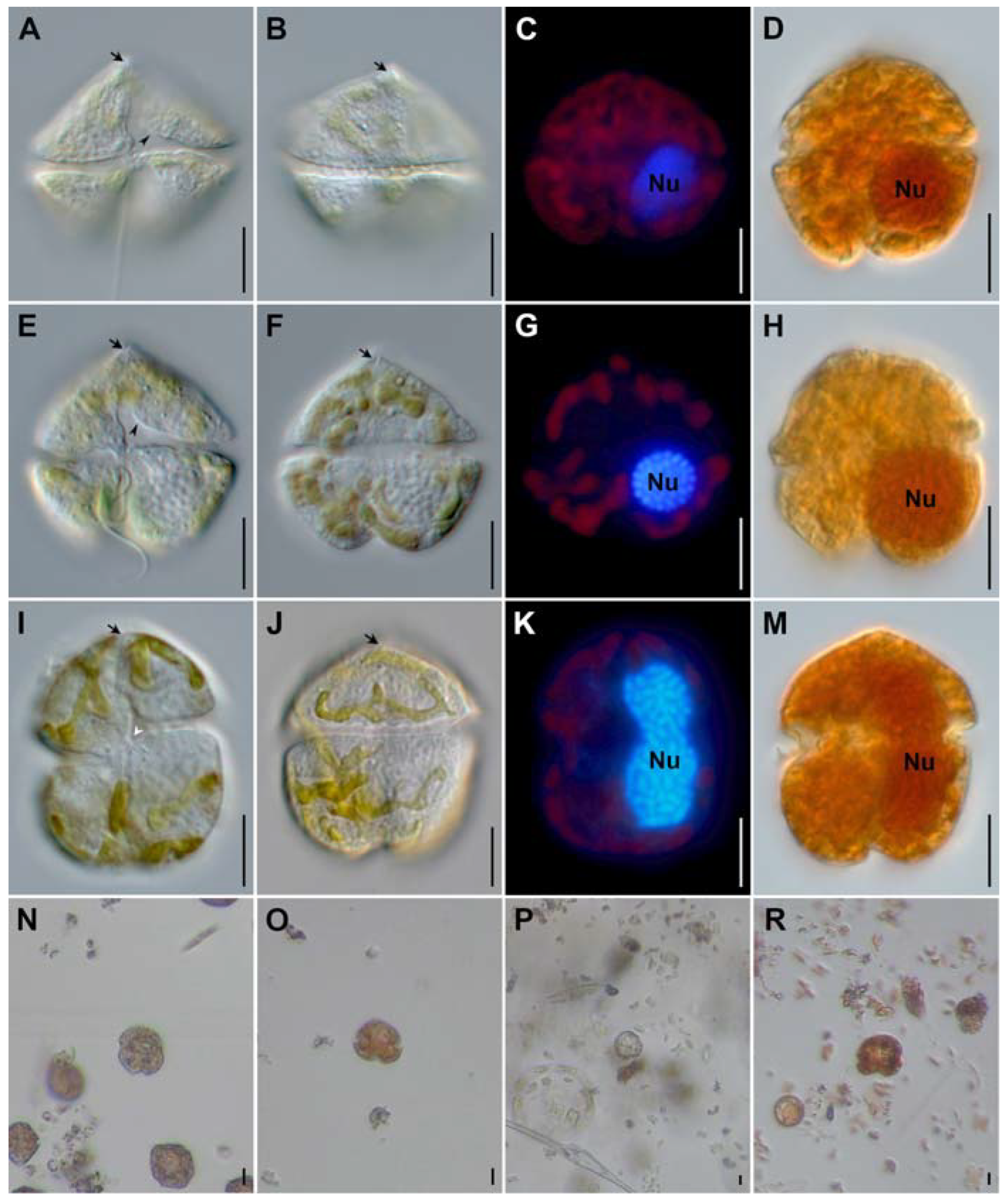
Bovine anaplasmosis is a tick-borne disease caused by an obligate intercellular Gram-negative bacterium named Anaplasma (A.) marginale. In this study, we report the seasonal prevalence, potentially associated risk factors and phylogeny of A. marginale in cattle of three different breeds from Multan District, Southern Punjab, Pakistan. A total of 1020 blood samples (crossbred, n = 340; Holstein Friesian, n = 340; and Sahiwal breed, n = 340) from apparently healthy cattle were collected on a seasonal basis from March 2020 to April 2021. Based on PCR amplification of the msp5 partial sequence, overall, the A. marginale prevalence rate was estimated at 11.1% (113/1020) of the analyzed cattle samples. According to seasons, the highest prevalence rate was observed in autumn (16.5%), followed by winter (10.6%) and summer (9.8%), and the lowest was recorded in the spring (7.5%). The crossbred and Sahiwal cattle were the most susceptible to A. marginale infection, followed by Holstein Friesian cattle (7.9%). Analysis of epidemiological factors revealed that cattle reared on farms where dairy animals have tick loads, dogs coinhabit with cattle and dogs have tick loads have a higher risk of being infected with A. marginale. In addition, it was observed that white blood cell, lymphocyte (%), monocyte (%), hematocrit, mean corpuscular hemoglobin and mean corpuscular hemoglobin concentrations were significantly disturbed in A. marginale-positive cattle compared with non-infested cattle. Genetic analysis of nucleotide sequences and a phylogenetic study based on msp5 partial sequencing demonstrated that this gene appears to be highly conserved among our isolates and those infecting apparently healthy cattle from geographically diverse worldwide regions. The presented data are crucial for estimating the risk of bovine anaplasmosis in order to develop integrated control policies against bovine anaplasmosis and other tick-borne diseases infecting cattle in the country.
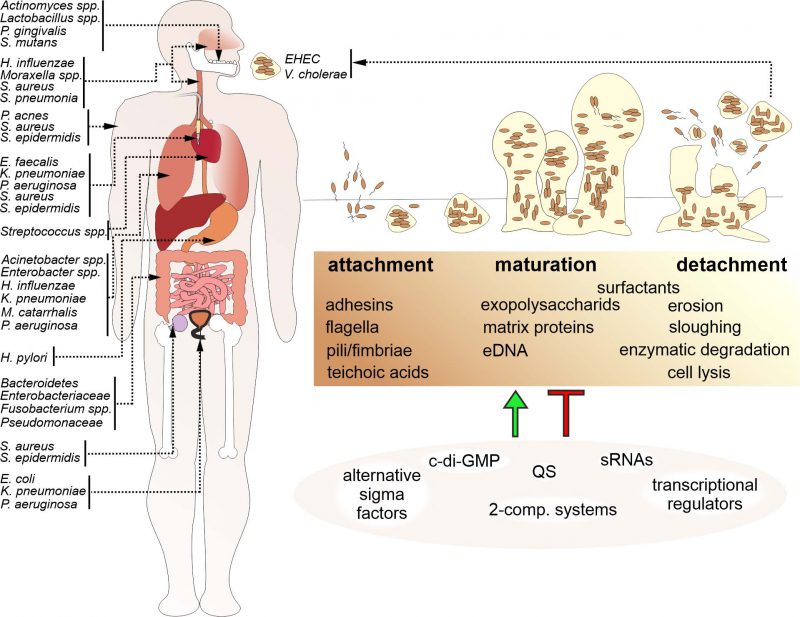
Biofilms by bacterial human pathogens: Clinical relevance

Bacterial Fish Pathogens: Disease of Farmed and Wild Fish

Bloodborne Pathogens Certificate Templates 6 FREE Designs

Pathogens, Free Full-Text

Sequential Infection with Common Pathogens Promotes Human-like
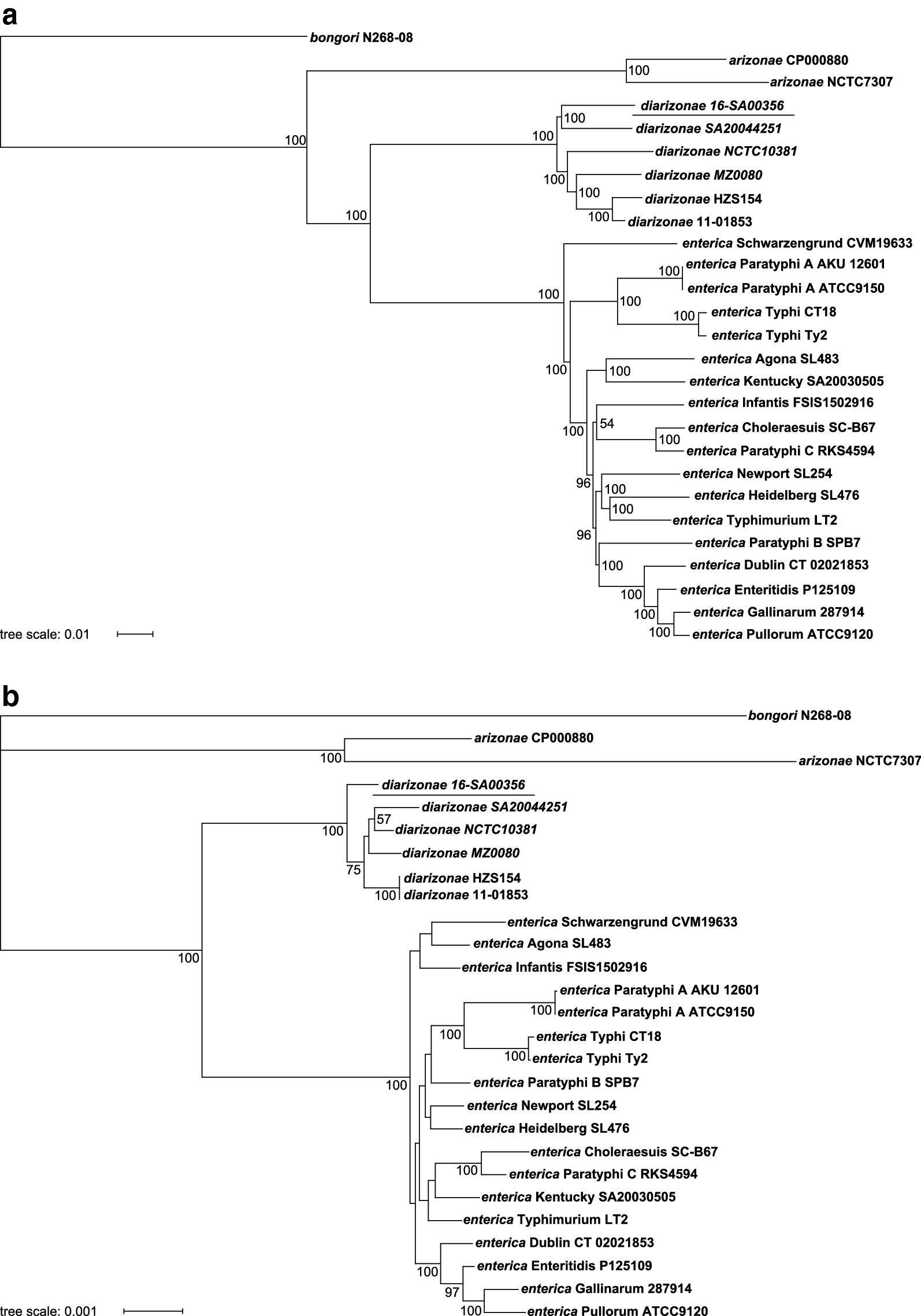
First complete genome sequence and comparative analysis of
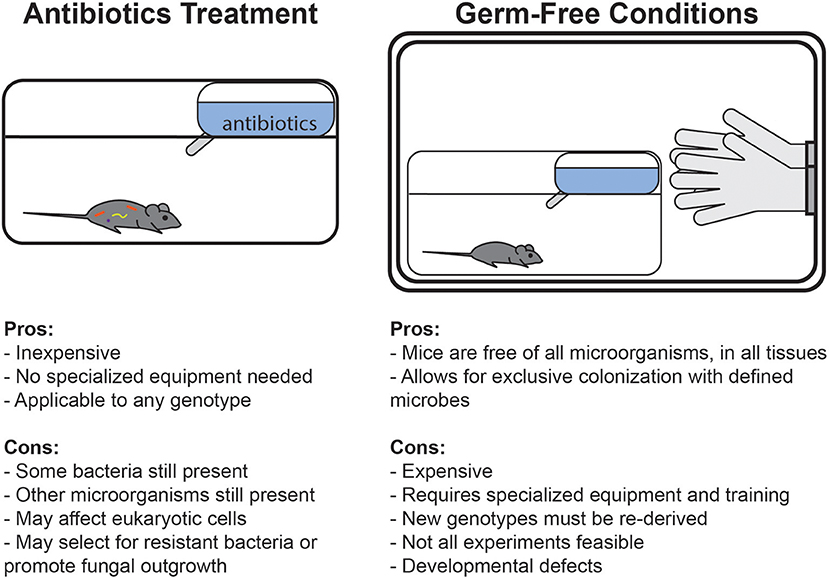
Frontiers Mouse Microbiota Models: Comparing Germ-Free Mice and
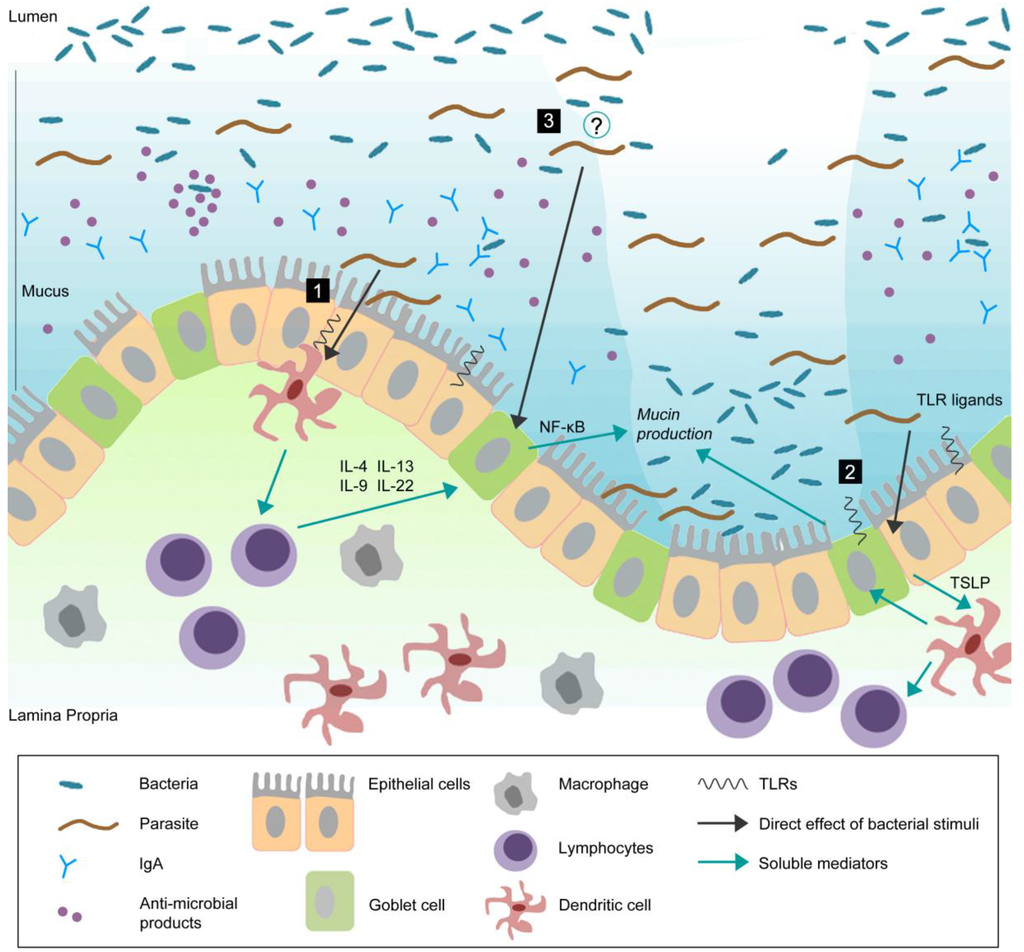
Pathogens, Free Full-Text
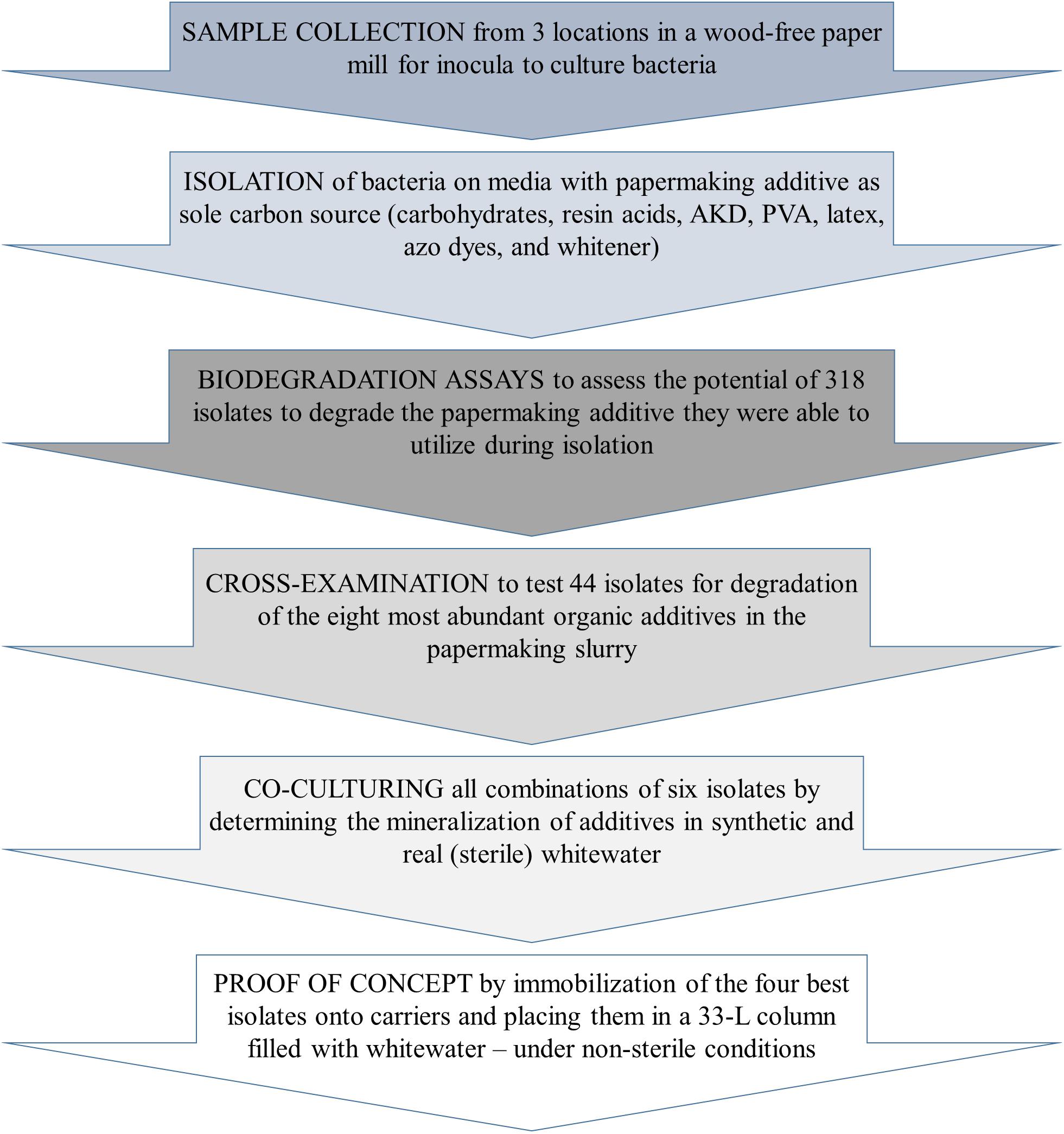
Frontiers Isolation, Identification, and Selection of Bacteria
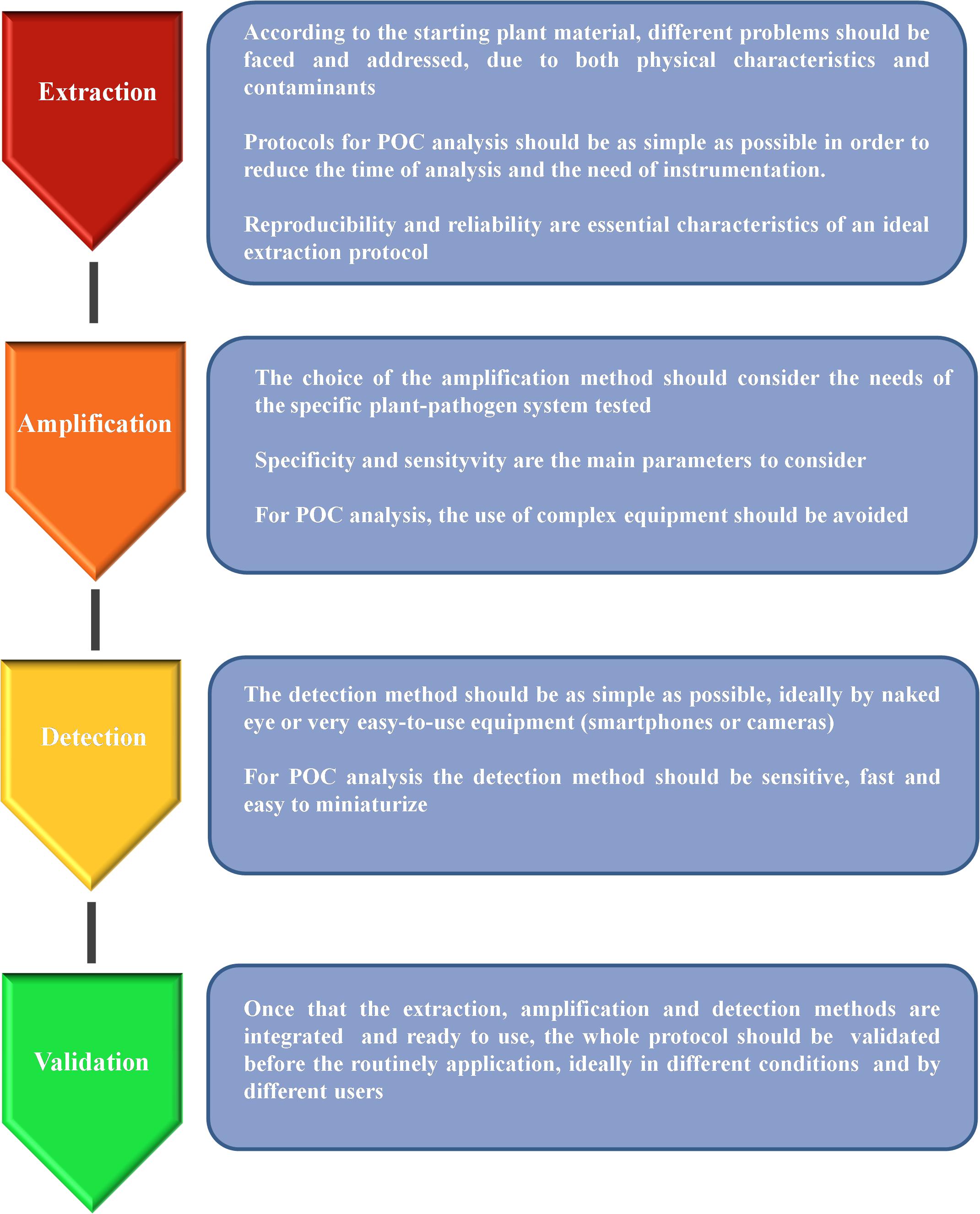
Frontiers Molecular Approaches for Low-Cost Point-of-Care
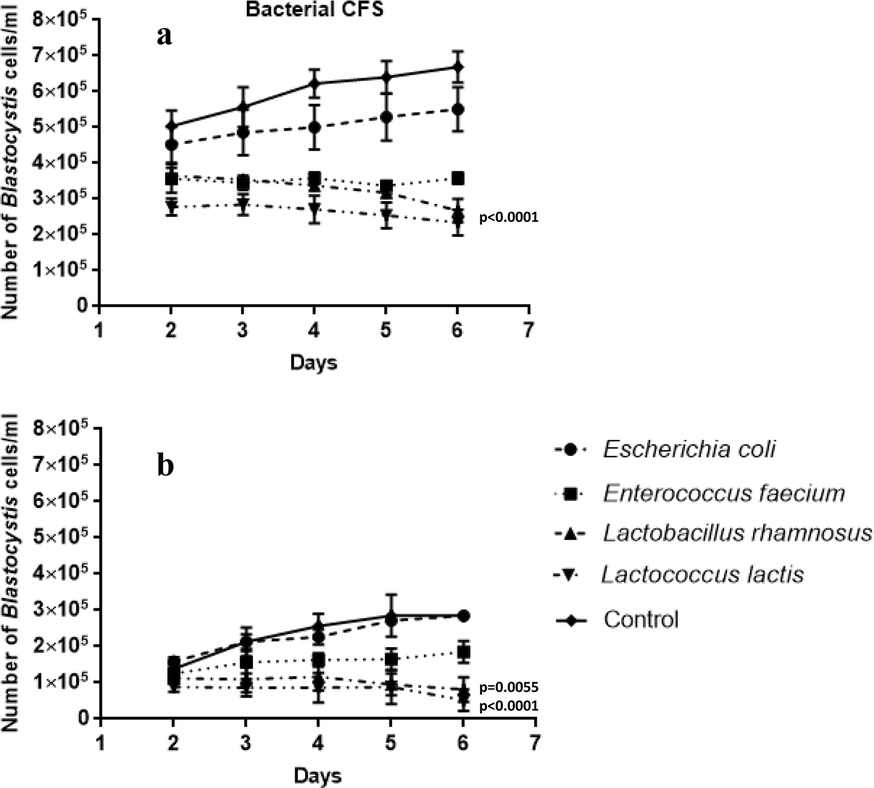
The influence of probiotic bacteria and human gut microorganisms
de
por adulto (o preço varia de acordo com o tamanho do grupo)